3.2 Parallel Runs
Some of the TURBOMOLE modules are parallelized using the message passing interface
(MPI) for distributed and shared memory machines or with OpenMP or multi-threaded
techniques for shared memory and multi-core machines.
Generally there are two hardware scenarios which determine the kind of parallelization
that is possible to use:
The list of programs parallelized includes presently:
- ridft — parallel ground state Hartree-Fock and DFT energies including RI-J
and the multipole accelerated RI (MA-RI-J), SMP and MPI
- rdgrad — parallel ground state gradients from ridft calculations, SMP and
MPI
- dscf — Hartree-Fock and DFT ground state calculations for all available DFT
functionals, without the usage of RI-J approximation, SMP and MPI
- riper — parallel ground state DFT energies for molecular and periodic
systems. SMP only.
- grad — parallel ground state gradients from dscf calculations, SMP and MPI
- ricc2 — parallel ground and excited state calculations of energies and
gradients at MP2 and CC2 level using RI, as well as energy calculations of
other wave function models, see chapter 10.6. SMP and MPI
- ccsdf12 — parallel ground state energies beyond MP2/CC2 and excitation
energies beyond CC2. SMP
- pnoccsd — parallel ground state energies at the OSV-PNO-MP2 and
OSV-PNO-MP2-F12 level, SMP and MPI
- mpgrad — parallel conventional (i.e. non-RI) MP2 energy and gradient
calculations. Please note that RI-MP2 is one to two orders of magnitude faster
than conventional MP2, so even serial RI-MP2 will be faster than parallel MP2
calculations. SMP and MPI
- aoforce — parallel Hartree-Fock and DFT analytic 2nd derivatives for
vibrational frequencies, IR spectra, generation of Hessian for transition state
searches and check for minimum structures. SMP and MPI
- escf — parallel TDDFT, RPA, CIS excited state calculations (UV-Vis and
CD spectra, polarizabilities). SMP only.
- egrad — parallel TDDFT, RPA, CIS excited state analytic gradients,
including polarizability derivatives for RAMAN spectra. SMP only.
- NumForce — this script can used for a trivial parallelization of the numerical
displaced coordinates. SMP and MPI
Additional keywords necessary for parallel runs with the MPI binaries are not needed.
When using the parallel version of TURBOMOLE, scripts are replacing the binaries. Those
scripts prepare a usual input, run the necessary steps and automatically start the
parallel programs. The users just have to set environment variables, see Sec. 3.2.1
below.
To use the OpenMP parallelization only an environment variable needs to be set. But to
use this parallelization efficiently one should consider a few additional points, e.g. memory
usage, which are described in Sec. 3.2.2.
3.2.1 Running Parallel Jobs — MPI case
The parallel version of TURBOMOLE runs on all supported systems:
- workstation cluster with Ethernet, Infiniband, Myrinet (or other) connection
- SMP systems
- or combinations of SMP and cluster
Setting up the parallel MPI environment
In addition to the installation steps described in Section 2 (see page 48) you just have to
set the variable PARA_ARCH to MPI, i.e. in sh/bash/ksh syntax:
This will cause sysname to append the string _mpi to the system name and the
scripts like jobex will take the parallel binaries by default. To call the parallel
versions of the programs ridft, rdgrad, dscf, grad, ricc2, or mpgrad from your
command line without explicit path, expand your $PATH environment variable
to:
export PATH=$TURBODIR/bin/‘sysname‘:$PATH
The usual binaries are replaced now by scripts that prepare the input for a parallel run
and start mpirun (or poe on IBM) automatically. The number of CPUs that shall be used
can be chosen by setting the environment variable PARNODES:
The default for PARNODES is 2.
Finally the user can set a default scratch directory that must be available on all nodes.
Writing scratch files to local directories is highly recommended, otherwise the scratch files
will be written over the network to the same directory where the input is located. The
path to the local disk can be set with
export TURBOTMPDIR=/scratch/username
This setting is automatically recognized by most parallel programs. Note:
- This does not set the path for the integral scratch files for dscf (see section
below about twoint of keyword $scfintunit).
- In MPI parallel runs the programs attach to the name given in $TURBOTMPDIR
node-specific extension (e.g. /scratch/username-001) to avoid clashes
between processes that access the same file system. The jobs must have
the permissions to create these directories. Therefore one must not set
$TURBOTMPDIR to something like /scratch which would result in directory
names like
/scratch-001 — which usually can not be created by jobs running under a
standard user account.
So please set the temporary directory for parallel files to a local
file system at a position you are allowed to generate directories, like
/tmp/myname/tm-jobs/mpifiles
MPI versions, distributions and flavours
TURBOMOLE is using the MPI version which has been utilized to generate the binaries. To
make sure that the parallel version is running, no matter which MPI flavour you have
installed on your machines, TURBOMOLE does include the run-time version of the MPI
flavour it needs.
____________________________________________________________________________ Please
do not try to use TURBOMOLE with your local MPI version (OpenMPI, MPICH, ...)! Do
not call the parallel MPI binaries directly, just set $PARA_ARCH as described
above and call the modules the same way you use them in the serial version.
______
On Linux for PCs and Windows systems either IBM Platform MPI (formerly known as
HP-MPI, now also known as IBM Spectrum MPI) is used and included — see IBM
Platform MPI or Intel MPI Intel MPI.
COSMOlogic ships TURBOMOLE with a IBM Platform MPI Community Edition or
the full Intel MPI version. TURBOMOLE users do not have to install or license
IBM Platform MPI or Intel MPI themselves. Parallel binaries will run out of the
box on the fastest interconnect that is found - Infiniband, Myrinet, TCP/IP,
etc.
Note: most parallel TURBOMOLE modules need an extra server running in addition to the
clients. This server is included in the parallel binaries and it will be started automatically
— but this results in one additional task that usually does not need any CPU time. So if
you are setting PARNODES to N, N+1 tasks will be started.
If you are using a queuing system or if you give a list of hosts where TURBOMOLE jobs shall
run on (see below), make sure that the number of supplied nodes match $PARNODES — e.g.
if you are using 4 CPUs via a queuing system, make sure that $PARNODES is set to
4.
Starting parallel jobs
After setting up the parallel environment as described in the previous section, parallel jobs
can be started just like the serial ones. If the input is a serial one, it will be prepared
automatically for the parallel run.
For the additional mandatory or optional input for parallel runs with the ricc2 program
see Section 10.6.
Running calculations on different nodes
If TURBOMOLE is supposed to run on a cluster, we highly recommend the usage of a
queuing system like PBS, Univa/SGE GridEngine or LFS. The parallel version of
TURBOMOLE will automatically recognise that it is started from within one of the queuing
systems:
- PBS (Torque/Maui)
- LSF
- SLURM
- SGE or Univa Grid Engine
and the binaries will run on the machines those queuing systems provide.
Important: Make sure that the input files are located on a network directory like an NFS
disk which can be accessed on all nodes that participate at the calculation.
If parallel jobs are started outside a queuing system, or if you have a non-supported or a
non-default installation of above mentioned queuing systems, the number of nodes and
their names can also be provided by the user. A file that contains a list of machines has to
be created, each line containing one machine name:
node1
node1
node2
node3
node4
node4
And the environment variable $HOSTS_FILE has to be set to that file:
export HOSTS_FILE=/nfshome/username/hostsfile
Note: Do not forget to set $PARNODES to the number of lines in $HOSTS_FILE, unless you
have set in addition OMP_NUM_THREADS (see below).
Note: In general the stack size limit has to be raised to a reasonable amount
of the memory (or to unlimited). In the serial version the user can set this by
ulimit -s unlimited on bash/sh/ksh shells or limit stacksize unlimited on csh/tcsh
shells. However, for the parallel version that is not sufficient if several nodes are used, and
the /etc/security/limits.conf files on all nodes might have to be changed. See chapter
2.2 of this documentation, page 56
OpenMP/MPI hybrid version
Some TURBOMOLE modules like dscf, grad, aoforce, ricc2 or pnoccsd are parallelized
using a hybrid OpenMP/MPI scheme. For those modules it is sufficient to start just one
single process per node. In addition, please set
export OMP_NUM_THREADS=<number of cores per node>
when starting the job. This environment variable will be exported to each node
such that the processes started there will open <number of cores per node>
threads.
Memory for parallel jobs
Since there are several different parallel versions of the individual TURBOMOLE modules
available, the meaning of the keywords to set memory ($ricore and $maxcor) can be
quite confusing. A lot of problems can be avoided if following points are taken care
of:
- for almost all cases increasing $ricore will not speed up the calculation but
increase memory consumption significantly. It is therefore recommended to set
$ricore to a small value like 100 or 500. Except:
- usage of RI-JK does benefit from large $ricore values. Check if $rik is present
in your control file — and if yes, try to increase the memory to a value which
your machines are capable to handle.
- many post-SCF programs read and use $maxcor. There are a couple of options
to this keyword like per_proc to define the amount of memory per process or
per_node for memory settings on one node, etc. A detailed description can be
found in chapter 20.2.2 on page 705.
- if you worry about speed in RI-DFT calculations, make sure to have $marij
in your control file
Testing the parallel binaries
The binaries ridft, rdgrad, dscf, grad, and ricc2 can be tested by the usual test suite:
go to $TURBODIR/TURBOTEST and call TTEST
Note: Some of the tests are very small and will only pass properly if 2 CPUs are used at
maximum. Therefore TTEST will not run any test if $PARNODES is set to a higher value than
2.
If you want to run some of the larger tests with more CPUs, you have to edit the DEFCRIT
file in TURBOMOLE/TURBOTEST and change the $defmaxnodes option.
Linear Algebra Settings
The number of CPUs and the algorithm of the linear algebra part of Turbomole depends
on the settings of $parallel_platform:
-
cluster
- – for clusters with TCP/IP interconnect. Communication is avoided by using
an algorithm that includes only one or few CPUs.
-
MPP
- – for clusters with fast interconnect like Infiniband or Myrinet. Number of
CPUs that take part at the calculation of the linear algebra routines depends
on the size of the input and the number of nodes that are used.
-
SMP
- – all CPUs are used and SCALapack (see http://www.netlib.org/scalapack/)
routines are involved.
The scripts in $TURBODIR/mpirun_scripts automatically set this keyword depending on
the output of sysname. All options can be used on all systems, but especially the SMP
setting can slow down the calculation if used on a cluster with high latency or small
bandwidth.
Sample simple PBS start script
#!/bin/sh
# Name of your run :
#PBS -N turbomole
#
# Number of nodes to run on:
#PBS -l nodes=4
#
# Export environment:
#PBS -V
# Set your TURBOMOLE pathes:
######## ENTER YOUR TURBOMOLE INSTALLATION PATH HERE ##########
export TURBODIR=/whereis/TURBOMOLE
###############################################################
export PATH=$TURBODIR/scripts:$PATH
## set locale to C
unset LANG
unset LC_CTYPE
# set stack size limit to unlimited:
ulimit -s unlimited
# Count the number of nodes
PBS_L_NODENUMBER=‘wc -l < $PBS_NODEFILE‘
# Check if this is a parallel job
if [ $PBS_L_NODENUMBER -gt 1 ]; then
##### Parallel job
# Set environment variables for a MPI job
export PARA_ARCH=MPI
export PATH="${TURBODIR}/bin/‘sysname‘:${PATH}"
export PARNODES=‘expr $PBS_L_NODENUMBER‘
else
##### Sequentiel job
# set the PATH for Turbomole calculations
export PATH="${TURBODIR}/bin/‘sysname‘:${PATH}"
fi
#VERY important is to tell PBS to change directory to where
# the input files are:
cd $PBS_O_WORKDIR
######## ENTER YOUR JOB HERE ##################################
jobex -ri > jobex.out
###############################################################
3.2.2 Running Parallel Jobs — SMP case
The SMP version of TURBOMOLE currently combines three different parallelization schemes
which all use shared memory:
- dscf, grad, aoforce, escf, egrad, ricc2, ccsdf12, pnoccsd, evib, odft,
rirpa and riper are partially parallelized with OpenMP for applications on
shared-memory, in particular multi-CPU and multi-core, machines.
- aoforce, escf, egrad, ridft and rdgrad are also parallelized as described
in [26]
- ridft and rdgrad are parallelized with MPI using shared memory on SMP
systems. This is also the default version for SMP systems, not just for MPI
runs.
Setting up the parallel SMP environment
In addition to the installation steps described in Section 2 (see page 48) you just have to
set the variable PARA_ARCH to SMP, i.e. in sh/bash/ksh syntax:
This will cause sysname to append the string _smp to the system name and the scripts like
jobex will take the parallel binaries by default. To call the parallel versions of the
programs (like ridft or aoforce) from your command line without explicit path, expand
your $PATH environment variable to:
export PATH=$TURBODIR/bin/‘sysname‘:$PATH
The usual binaries are replaced now by scripts that prepare the input for a parallel run
and start the job automatically. The number of CPUs that shall be used can be chosen by
setting the environment variable PARNODES:
The default for PARNODES is 2.
NOTE: Depending on what you are going to run, some care has to be taken that the
system settings like memory limits, etc. will not prevent the parallel versions to run. See
the following sections.
OpenMP parallelization of almost all time consuming modules
The OpenMP parallelization does not need any special program startup. The binaries can
be invoked in exactly the same manner as for sequential (non-parallel) calculations. Just
set the environment variable PARNODES to the number or threads that should be used by
the programs. The scripts will set OMP_NUM_THREADS to the same value and start the
OpenMP binaries directly. The number of threads is essentially the max. number of CPU
cores the program will try to utilize. To exploit e.g. all eight cores of a machine with two
quad-core CPUs set
(for csh and tcsh use setenv PARNODES=8).
Presently the OpenMP parallelization of ricc2 comprises all functionalities apart from the
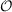 (
(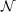 4)-scaling LT-SOS-RI functionalities (which are only parallelized with MPI) and
expectation values for Ŝ2 (not parallelized). Note that the memory specified with $maxcor
is for OpenMP-parallel calculation the maximum amount of memory that will be
dynamically allocated by all threads together. To use your computational resources
efficiently, it is recommended to set this value to about 75% of the physical memory
available for your calculations.
4)-scaling LT-SOS-RI functionalities (which are only parallelized with MPI) and
expectation values for Ŝ2 (not parallelized). Note that the memory specified with $maxcor
is for OpenMP-parallel calculation the maximum amount of memory that will be
dynamically allocated by all threads together. To use your computational resources
efficiently, it is recommended to set this value to about 75% of the physical memory
available for your calculations.
For Localized Hartree-Fock calculations please use the dscf program which is parallelized
using OpenMP. In this case an almost ideal speedup is obtained because the most
expensive part of the calculation is the evaluation of the Fock matrix and of the
Slater-potential, and both of them are well parallelized. The calculation of the
correction-term of the grid will use a single thread.
The OpenMP parallelization of riper covers all contributions to the Kohn-Sham matrix
and nuclear gradient. Hence an almost ideal speedup is obtained.
Restrictions:
- In the ricc2 program the parts related to RI-MP2-F12, LT-SOS-RI-MP2
or calculation of expectation values for Ŝ2 do not (yet) use OpenMP
parallelization. If the OpenMP parallelization is switched on (by setting
OMP_NUM_THREADS) these parts will still be executed sequentially.
- In the dscf program the $incore option will be ignored if more than one
thread is used. Semi-direct dscf calculations (i.e. if a size larger than 0 is given
two-electron integral scratch file in $scfintunit) can not be combined with
the OpenMP parallel runs. (The program will then stop with error message in
the first Fock matrix construction.)
Multi-thread parallelization of dscf, grad, aoforce, escf, egrad, ridft and
rdgrad
The parallelization of those modules is described in [26] and is based on fork() and Unix
sockets. Except setting PARNODES which triggers the environment variable SMPCPUS, the
environment variable
has to be set. Alternatively, the binaries can be called with -smpcpus <N> command line
option or with the keyword $smp_cpus in the control file.
The efficiency of the parallelization is usually similar to the default version, but for
ridft and rdgrad RI-K is not parallelized. If density convergence criteria ($denconv) is
switched on using ridft and if no RI-K is being used, the multi-threaded version should
be used.
SMP/MPI version of ridft and rdgrad
Since TURBOMOLE version 7.2 the usage of GlobalArrays has been omitted. Instead, a set of
routines which utilize shared memory on a node has been implemented. Both modules,
ridft and rdgrad, start each process as an individual MPI instance. Processes on the
same node are then collected to collectively store and use data in a shared memory region.
This avoids excessive memory usage and reduces the amount of memory requirements
significantly, especially compared to the old MPI implementation (which has been
used by default in former TURBOMOLE versions). It is nevertheless recommended
to
- run jobs in parallel only if the molecules and/or basis set size is large enough
— several hundred basis functions for non-hybrid functionals, or few hundred
for hybrid functional calculations. The RI approximation in combination with
MARI-J is already very fast in the serial version, usage of many cores would
introduce a communication overhead which exceeds the computational time on
a single core.
- not ask for too much memory. For medium sized to large molecules adding a
significant amount of $ricore will not speed up the calculation, but eventually
lead to overstressed systems (or even memory swapping) or failure of jobs due
to too large memory requirements. So please do not set large $ricore values
in such cases, a few ten or hundred MB are sufficient (even zero works equally
well).
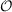 (
(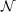 4)-scaling LT-SOS-RI functionalities (which are only parallelized with MPI) and
expectation values for Ŝ2 (not parallelized). Note that the memory specified with $maxcor
is for OpenMP-parallel calculation the maximum amount of memory that will be
dynamically allocated by all threads together. To use your computational resources
efficiently, it is recommended to set this value to about 75% of the physical memory
available for your calculations.
4)-scaling LT-SOS-RI functionalities (which are only parallelized with MPI) and
expectation values for Ŝ2 (not parallelized). Note that the memory specified with $maxcor
is for OpenMP-parallel calculation the maximum amount of memory that will be
dynamically allocated by all threads together. To use your computational resources
efficiently, it is recommended to set this value to about 75% of the physical memory
available for your calculations.