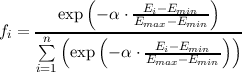
The reliable structure prediction of gas-phase clusters is complicated by a large number of possible structural isomers, often in combination with several low-lying electronic states. Brut force determination of the most stable configuration by manual construction of all models and following local structure optimizations is a formidable task. Instead, the DoDo program allows the automatic determination of the most stable molecular structures using the genetic algorithm (GA) to locate the global minimum of the total (electronic) energy. The GA is a search heuristic that employs principal mechanisms of natural evolution such as inheritance, mutation, selection, and crossover for exploration of the potential energy surface.
Generally, in GA a population of candidate structures is evolved toward improved solutions. Within the DoDo program the GA is initialized with a pool of randomly (automatically) generated structures, with initial structure models supplied by the user – so called seeds – or a combination of both. Every seed is used one or more times to create initial structures by randomly adding or removing atoms until the predefined composition and system size is reached. All initial structures are subsequently geometry optimized to the nearest local minimum. Next, two evolutionary operators – crossover and mutation – are used to exchange structure information between the members of the current population. In the crossover, pairs of structures are chosen to act as parents that will produce a new structure for the next generation. Within each parent pair, random pieces are exchanged to form the new mixed structure, the child, which is subsequently optimized to the nearest local minimum as well. The assumption is that the child will combine the good structural features of the parents and thus will be more stable than either one. Evolutionary pressure towards improved children is added to bias the search in the right direction. This is achieved by selecting parents that have a relatively high fitness that is defined as a function of their total energy:
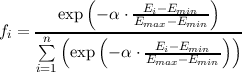 | (5.5) |
where fi is the fitness of the ith individual, n is the number of individuals, α is a constant scaling factor, and Ei is the energy of that individual relative to the maximum (Emax) and minimum (Emin) energies of structures in the whole population. Since the child structures not always present a better fitness than their parents, elitism or natural selection is applied. It is achieved by simply replacing parents with a worse fitness by children with a better one in the structures pool. In order to maintain a maximum diversity during the GA runs similarity recognition is used that allows only distinct structures to be included in the pool. To prevent trapping of the population in local minima mutation is added in which random changes are introduced to randomly chosen structures in the pool.
The determination of the global minimum structure typically requires, depending on the system size, several hundreds to few thousands of local structure optimizations of the children and mutated structures. Therefore, the DoDo program uses a simple parallelization in which a predefined number (usually 10-100, depending on computer resources) of local optimizations are performed simultaneously, each preferably running on a single or just a few CPU cores. When a number of local optimizations finish, i.e. the number of running/queued geometry optimizations falls below a minimum value a, natural selection is applied and new child structures are generated by crossover or mutation and submitted for local optimization until the maximum number b of simultaneously running/queued optimizations is reached. This pool-based GA allows for an optimal utilization of computer resources since the algorithm is not required to wait for the completion of local optimizations of all children structures in a particular generation. The progress of the pool-based GA is then measured by number of locally optimized structures rather than by the generation count as in case of the generation-based GA (a = 1).
More details of the implemented GA and application examples can be found in Ref. [49] and references therein.
In order to start a global optimization procedure one directory is required containing at least:
tmole20 -l -c coord turbo.in > turbo.log &> turbo.err
A file containing seed structures can be optionally included in the directory. The GA is started by calling the DoDo main program with:
nohup genetic --gen_inp=INPUT_FILE >& OUTPUT_FILE &
This initiates a process running in background of the clusters front-end that: i) supervises the GA run, ii) stores the population and the history of the run, iii) performs the natural selection algorithm, iv) calls specialized genetic operator modules, v) performs input/output operations, vi) interacts with the queueing system. The generated output file summarizes the progress of the run including information about minimal and average energies of structures in a certain population, crossover and mutation operations as well as the status of queued jobs.
Each local optimization is performed in a separate directory and as an independent job (stored in “calculations”). The Tmole input file and job script supplied by the user are copied into the created working directory. Then, the molecular structure to be optimized is automatically exported as coord file and the job is submitted to the queuing system using the command compatible with the given job scheduling software (e.g. qsub for PBS). The DoDo package stores the unique identifier returned by the queuing system upon the job submission. Job progress is periodically checked using the identifier and the proper command (e.g. qstat for PBS). After the job is finished, the relevant output files created by TURBOMOLE are parsed in order to obtain the final structure definition and the corresponding energy.
The structures of the current population (with the highest fitness) are stored in a file named “POPULATION.dodo” while the file “DEAD.dodo” contains all structures that were removed from population due to the natural selection algorithm. A complete history of previous populations and rejected structure models is stored in the directory “structures”. These files as well as the files for the initial seed structures use the internal DoDo format (distance units: Bohr).
For conversion between the DoDo format and other commonly used file formats (e.g., car and xyz) one can employ the script convert along with the following options:
name of file to convert (default: POPULATION.dodo)
file format of base structure: car, xyz or dodo (default: dodo)
output file format: car, arc, xyz, mxyz (xyz file optimized for molden) or dodo (default: mxyz)
change coordinates from ┼ngstrom to Bohr (default: False)
change coordinates from Bohr to ┼ngstrom (default: False)
Should it be necessary to abort a GA run one can use genetic --clear to remove all submitted jobs from the queue before stopping the main process. In order to restart the GA, set the number of (random) initial structures to zero and remove the declaration of the seed file (if used) in the DoDo input file. Then, the latest “POPULATION.dodo” and “DEAD.dodo” files from the previous run are automatically read when genetic is executed again.
A sample input file used for the DoDo genetic algorithm, “genetic_example.inp”, is generated by calling:
genetic --tmole_example
Such an input file presents all keywords implemented in the program. For clarity, these keywords are separated in sections but the file is (almost) free formatted. The hash sign # starts a comment. The keywords and corresponding default values are summarized in the list presented below.
Defines the maximum population size. SIZE > 1, default: SIZE = 20.
Defines the minimum and maximum number of the local optimization jobs
present at any moment in the queue. Use MIN = 1 to have a ’standard’
generation-based GA run. MAX ≥ MIN ≥ 1, defaults: MIN = 1, MAX = 10.
Defines the total number of structures optimized during the run. Default: MAX
= 100.
Initial structures generator settings
Defines the maximum number of tries performed in order to generate a single
initial structure model. Default: MAX = 1000.
Defines the total number of generated initial structures. Default: NUMBER =
20.
Defines the multipliers for atomic radii used to check whether two atoms are
bound. Defaults: A = 0.7, B = 1.2.
The composition block defines the desired atomic composition of the initial
structures. SYMBOL is a chemical symbol of the given element. MIN and
MAX define minimum and maximum number of atoms for that element. Only
systems with constant composition during the GA run are currently available
(MAX = MIN).
Declares the file that contains seeds given in the internal DoDo format. Default:
FILENAME = OLD_POPULATION.dodo.
Defines how many initial species will be generated from each seed given in
the seed file. The number of the values has to be the same as the count of
the seeds given in the seed file, i.e. each seed in the seed file has to be
accounted for.
Crossover and mutation settings
This line is a sanity check. If this line is present in the input file, MIN has to
be equal to MAX for each SYMBOL in the composition section. Otherwise,
an error is raised. Only this mode is currently available.
Defines the maximum number of tries performed in order to generate a single
structure model. Default: MAX = 1000.
Defines the scaling factor α used in Eq. (5.5). Default: FACTOR = 1.0.
Defines the mutation probability of structures within the GA population.
Default: VALUE = 0.01.
Defines the maximum root mean square of the distances between the
corresponding atoms for which two structures are considered similar. Default:
VALUE = 0.3.
Defines the maximum distance [┼] used to seek for the corresponding atoms.
Default: VALUE = 0.3.
Defines which job scheduling software interface should be used. NAME = pbs
or NAME = hlrn, default: NAME = pbs.
Defines how often [seconds] the GA checks the queuing system for the presence
of submitted optimization jobs. Default: VALUE = 600.
Defines the computational platform (TURBOMOLE in combination with Tmole).
NAME = turbomole.
Defines the name of file which will be submitted to the queueing system.
Default: NAME = job.run.
This file is used by Tmole2.0. NAME given here HAS to agree with data given
in job file.